
Integrated
biochips debut
By
Eric Smalley,
Technology Research NewsWhen the computer chip was invented forty-four years ago, it set the stage for computers to shrink from room-size behemoths filled with light-bulb-size vacuum tubes to handheld devices powered by microscopic transistors.
Researchers from the California Institute of Technology are mirroring that effort with a chip that stores tiny drops of fluid rather than magnetic or electronic bits of information.
The researchers are aiming to replace roomfuls of chemistry equipment with devices based on a fluidic storage chip that can store 1,000 different substances in an area slightly larger than a postage stamp.
The technology could eventually allow experiments that involve hundreds or thousands of liquid samples to run on desktop or even handheld devices, potentially reducing the cost and complexity of medical testing, genetics research and drug development, said Stephen Quake, an associate professor of physics and applied physics at Caltech. "Small volumes mean lower cost for expensive reagents, and mean that samples can be tested for a broader range" of diseases, he said.
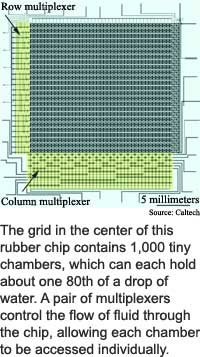
The fluidic storage chip has 1,000 chambers arranged in a 25 by 40 grid with 3,574 microvalves. "It's small plumbing -- pipes, valves, pumps, et cetera -- all integrated on a small rubber chip," said Quake.
Each chamber holds 250 picoliters, or about one 80th of a drop of water. The connecting channels are 100 microns wide and nine microns high, which is about twice as high as red blood cells are wide.
Like the bits that store 1s and 0s in computer memory, the chambers that store fluids at the intersections of the rows and columns of the researchers' chip can be accessed individually. The key is a pair of multiplexors that address each chamber by row and column. Computer memory chips use similar electronic multiplexors to access individual bits of digital information.
In the fluidic storage chip, the row multiplexor pushes fluids along one or more rows to fill or purge the chambers in those rows, and the column multiplexor applies pressure to close the input and output valves of the chambers along one or more of the columns.
The fluidics multiplexors allow the researchers to control the 1,000 chambers using only 22 connections to the chip, said Quake. "We can control exponentially many fluid lines," as outside connections, he said. Thirty connections could theoretically control 32,000 fluid lines and 40 connections could theoretically control one million fluid lines. "This greatly simplifies the input/output and connections required from the real world to the chip," he said.
To make the fluidic chip, the researchers etched patterns into plastic molds using the same photolithography process used to make computer chips, then used the molds to shape thin layers of rubber.
The top layer contains fluid channels and chambers, and the bottom layer holds multiplexors and control lines. In between is a thin sheet of rubber. The intersection of a fluid channel and a control line forms a valve; hydraulic pressure in the control line deflects the thin membrane between the top and bottom layers and pinches off the fluid channel. The multiplexors determine where the pressure is applied in order to control the flow.
The chip is made completely of flexible silicone rubber, rather than the hard silicon used in computer chips. Fluids enter the chip through steel pins connected through holes punched into the rubber, which forms a tight seal around the pins.
The researchers also made a chip-size comparator, which measures samples against a scale or standard to determine properties like the pH concentration of a fluid.
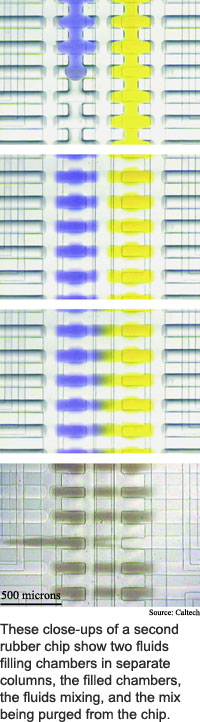
The researchers' comparator has an array of 256 chambers arranged in four columns of 64, and is about twice the size of the storage chip. It contains 2,056 microvalves and performs more complicated manipulations than the storage chip, according to Quake. Two fluids can be mixed in any number of the chambers and the results from any chamber in each column can be removed for further examination, he said.
The researchers took the comparator through its paces by loading individual bacteria into some of the chambers and adding a fluid that becomes fluorescent in the presence of a particular enzyme. This allowed the researchers to determine which bacteria produced the enzyme.
There are limitations to the rubber chips, according to Quake. Some liquids, like certain organic solvents, can break down the chip's rubber material, and there is a danger of contamination from molecules diffusing through the walls of the chambers and channels. There is also a possibility that some molecules will stick to the walls after the chip's contents have been emptied.
In addition, the chip designs are limited by the need to avoid cross-contamination as samples are shunted about. For example, the contents of only one of the 64 chambers in each column of the comparator can be removed without being contaminated because any residue from the first sample would contaminate subsequent samples passing through the channel, according to Quake.
The researchers' work is an impressive and significant advance, said Kenny Breuer, an associate professor of engineering at Brown University. "There have been many attempts at building such microfluidic elements, but this is by far the most complex that I have seen, and the approach... offers the most flexibility for building a wide variety of microfluidic systems," he said.
The system does have limitations, Breuer added. "There is... significant hidden machinery that is required to operate the device -- supplies of compressed air, banks of solenoidal valves and, most importantly, very large volumes of fluid that need to be flushed through the system as each cell is loaded and purged," he said. The volume of this supporting infrastructure could limit the size and complexity of fluidic systems made with this technology, he said.
It is also true, however, that the first electronic computer chips used large amounts of power and were not able to do much, but "still enabled a revolution in electronics and engineering," said Breuer. The ability to create large-scale integrated microfluidics systems with such complexity is very exciting, even if this particular design may eventually be supplanted by other approaches, he said.
The researchers next plan to use the devices in biological research, said Quake. "One area will be in environmental microbiology."
The technology could be used in practical applications in one to two years, said Quake. There are some manufacturing issues that need to be addressed, but "it is already working in some practical applications," he said. Quake is a director of Fluidigm Corporation, which is commercializing the technology.
Quake's research colleagues were Todd Thorsen and Sebastian Maerkl. They published the research in the September 26, 2002 online issue of the journal Science. The research was funded by the Defense Advanced Research Projects Agency (DARPA) and the Army Research Office (ARO).
Timeline: 1-2 years
Funding: Government
TRN Categories: Microfluidics and BioMEMS; Biotechnology; Engineering
Story Type: News
Related Elements: Technical paper, "Microfluidic Large-Scale Integration," Sciencexpress, September 26, 2002
Advertisements:
October 2/9, 2002
Page One
Integrated biochips debut
Metal mix boosts batteries
Plastic tag makes foolproof ID
Scheme hides Web access
Small jolts move artificial muscle
News:
Research News Roundup
Research Watch blog
Features:
View from the High Ground Q&A
How It Works
RSS Feeds:
News
Ad links:
Buy an ad link
| Advertisements:
|
 |
Ad links: Clear History
Buy an ad link
|
TRN
Newswire and Headline Feeds for Web sites
|
© Copyright Technology Research News, LLC 2000-2006. All rights reserved.