
DNA machines take a walk
By
Eric Smalley and Kimberly Patch,
Technology Research NewsResearchers working to form nanoscale machines and materials are increasingly tapping nature's building blocks. Two particularly helpful molecules are DNA, which encodes instructions for making the proteins that carry out life's processes, and the motor protein kinesin, which is part of the a cell's transportation system.
DNA molecules contain strings of four types of bases -- adenine, cytosine, guanine and thymine -- attached to a sugar-phosphate backbone. Single strands self-assemble into structures like life's familiar double helix when their bases match up. Researchers can manipulate artificial strands of DNA by controlling these connections.
Kinesin molecules have a pair of short extensions, or legs, at one end and a tail at the other end. The legs attach to a microtubule protein, and step to move the kinesin bipedal molecule forward along the microtubule molecule. The tail grabs cell structures like vesicles and mitochondria to transport the structures over a cell's extensive network of microtubules.
Several research teams have built DNA walkers, inspired in part by kinesin, that move along DNA tracks.
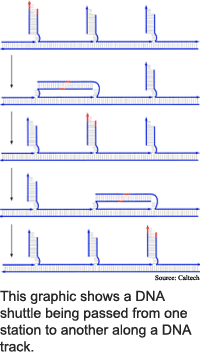
Researchers from Duke University and the University of Oxford in England have devised a a series of DNA stations that pass a DNA fragment from one to the next. The walker works autonomously, using enzymes present in the environment to initiate each step of the process.
Researchers at the California Institute of Technology have built a bipedal DNA walker that improves the gate of a walker originally designed by researchers at New York University from shuffling, with one leg always in back of the other, to leg-over-leg walking.
Both types of walkers could eventually be used to construct nanoscale devices, synthesize and deliver drugs, and carry out DNA computation. "Eventually, it may be possible to program synthetic motors that can haul diverse molecules along intricate paths for use in nanoscale factories or molecular medicine," said Niles Pierce, an assistant professor of applied and computational mathematics and bioengineering at Caltech.
The Caltech prototype is designed to mimic kinesin, and consists of a DNA track with four anchorage sites and a bipedal DNA walker. The anchorage sites are single strands of DNA set five nanometers apart along the double helix track, and the walker is a double helix structure with strands separated at one end.
The walker's loose strands form a pair of 23-base legs, and the four anchorage sites contain different sequences of 20 bases.
To set up the process, the researchers add an attachment strand that joins at one end with the first anchorage strand and at the other end with one leg. A short segment in the middle of the attachment strand that does not match up with the leg or anchorage strands provides flexibility.
To make the device take a step, the researchers add a second attachment strand that moves the second leg by fixing it the second anchorage strand. To make it take another step, the researchers free the first leg with a detachment strand that removes the first attachment strand, then swing the leg forward and bind it to the third anchorage with a third attachment strand. In this way, the walker progresses one leg in front of the other along the track.
The prototype falls short of kinesin in several regards, according to Pierce. Kinesin runs autonomously whereas the researchers' DNA walker requires DNA fuel strands -- the attachment and detachment strands -- to be administered at each step, he said. Kinesin also moves at about 100 steps per second while the researchers' device takes around two steps per hour, he said.
The Duke University walker consists of a double-stranded DNA track with three DNA anchorage site segments attached to the track by single-strand hinge segments of DNA. The anchorage DNA is double-stranded, but one strand shorter, leaving a three-base extension at the free end. The walker is a six-base single strand.
The walker begins attached to the first anchorage. The three bases at the free end of the walker attach to the end of the second anchorage, causing the first and second anchorages to swing on their hinges toward each other. An enzyme frees the walker from the first anchorage and alters the anchorage to keep the walker from stepping backwards. The free end of the walker then attaches to the third anchorage. A second enzyme cuts the walker free from the second anchorage to complete the move to the third anchorage site.
Unlike the bipedal DNA walkers, which require DNA strands to be added at each step of the process, the Duke device operates continuously because none of its components interfere with each other and so can all be present in the environment. "Our walker operates in an autonomous fashion while previous constructions by other groups require... the adding and removal of fuel DNA strands to drive the walker," said Hao Yan, a Duke University researcher who is now an assistant professor of chemistry and biochemistry at Arizona State University.
The DNA walker could eventually be used to carry out computations and to precisely transport nanoparticles of material, according to Yan. The walker can be programmed in several ways. "For example, we can encode information in [the] walker fragments as well as in the track so that while performing motion, the walker simultaneously carries out computation," said Yan.
The walker could also be programmed to transport tiny bits of material, said Yan. "If integrated with a well-defined large-scale nanostructure such as two-dimensional DNA nano-grids, the walker might be able to precisely transport a nanoparticle from one location to another location on the nanostructure... in a programmable and autonomous fashion," he said.
The DNA walkers advance DNA nanotechnology by anchoring controlled, progressive motion to a structure. This is a key step toward harnessing the work of DNA machines. "Ultimately, our objective in pursuing rational DNA design is to develop a molecular compiler that takes as input a conceptual design for a device and produces as output a list of DNA sequences that can be expected to assemble into the desired system," said Caltech's Pierce.
It is likely to take 10 years or more to design and build practical molecular motors from scratch, said Pierce. The Duke DNA walker could be used practically in five years, said Yan.
Yan's research colleagues were Peng Yin, Xiaoju G. Daniell, Andrew J. Turberfield and John H. Reif. Their work appeared in the Sept. 27, 2004 issue of Angewandte Chemie International Edition. The research was funded by the National Science Foundation (NSF).
Pierce's colleague was Jong-Shik Shin. Their work appeared in the July 14, 2004 issue of Nano Letters, and was funded by the Defense Advanced Research Projects Agency (DARPA), the Charles Lee Powell Foundation, the Ralph M. Parsons Foundation, and Caltech.
Timeline: 5 years; 10 years
Funding: Government
TRN Categories: DNA Technology; Nanotechnology
Story Type: News
Related Elements: Technical paper, "A Unidirectional DNA Walker That Moves Autonomously along a Track," Angewandte Chemie International Edition, September 27, 2004; technical paper, "A Synthetic DNA Walker for Molecular Transport," Nano Letters, July 14, 2004
Advertisements:
November 3/10, 2004
Page One
Ultrathin carbon speeds circuits
DNA machines take a walk
DNA in nanotubes sorts molecules
Single field shapes quantum bits
Briefs:
Nanotubes lengthen to centimeters
Coated nanotubes record light
Photonic crystal lasers juiced
Lasers move droplets
Molecules form nano containers
Square rings promise reliable MRAM
News:
Research News Roundup
Research Watch blog
Features:
View from the High Ground Q&A
How It Works
RSS Feeds:
News
Ad links:
Buy an ad link
| Advertisements:
|
 |
Ad links: Clear History
Buy an ad link
|
TRN
Newswire and Headline Feeds for Web sites
|
© Copyright Technology Research News, LLC 2000-2006. All rights reserved.